high-throughput protein dynamics
open-source software
Open-source software for complex sampling workflows on 2000+ GPUs
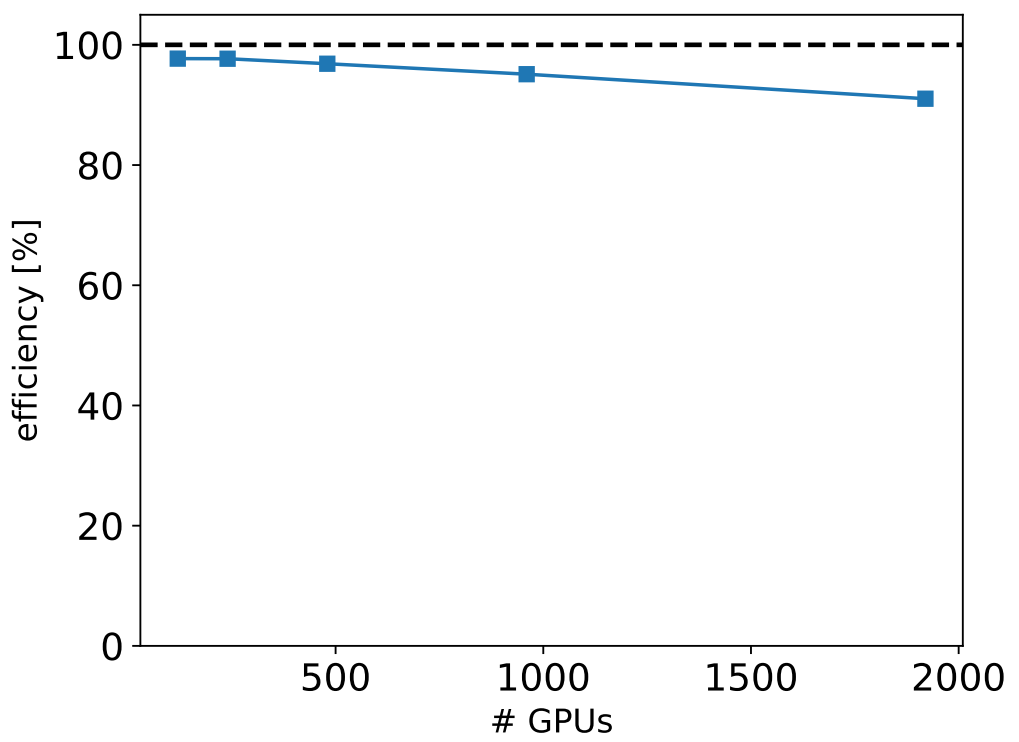
Running adaptive sampling strategies as described here requires efficient, scalable, and user-friendly execution of both the molecular dynamics and the iterative approach. I led the development of version 2 of the ExTASY software optimized for adaptive sampling. The code is available at https://github.com/ClementiGroup/ExTASY/. The modular python code allows an user-friendly swap of the molecular dynamics packages or the particular sampling methods with little code change. The Python-based interface to the interoperable and high-performance pilot-based run time system allows the scalability of ExTASY on supercomputers to reach 2000+ concurrent GPUs or concurrent molecular dynamics simulations.
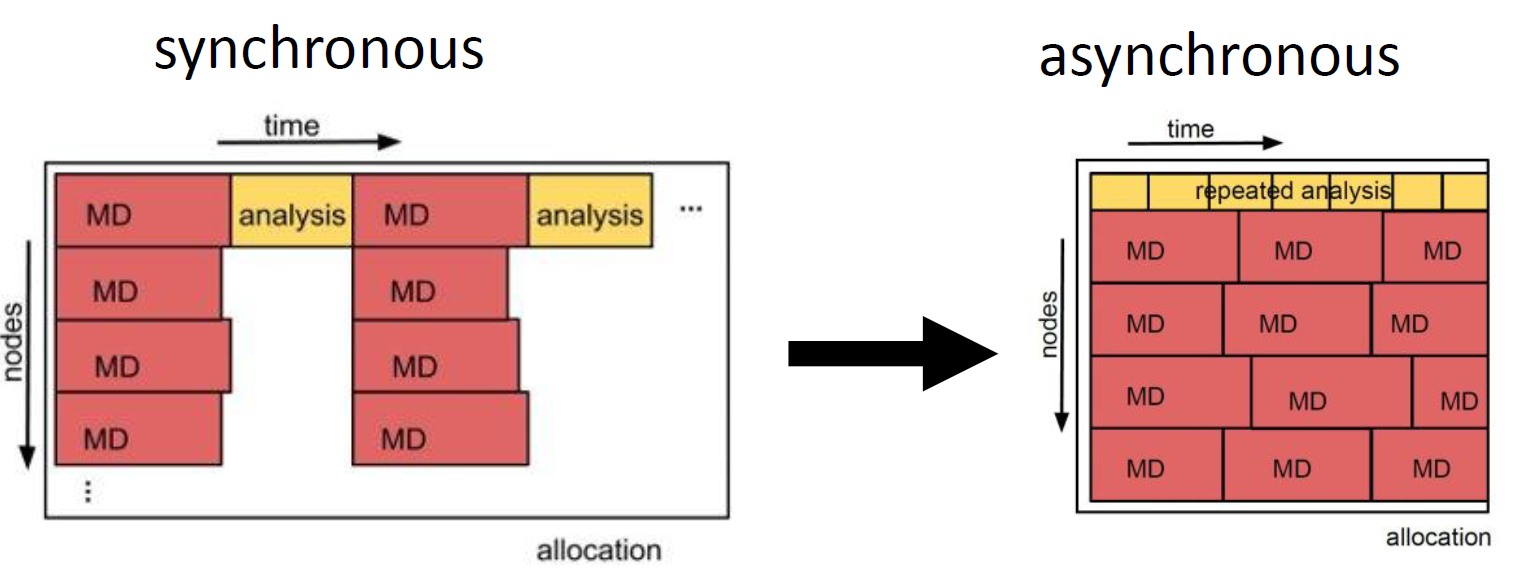
The high scalability of the ExTASY package is enabled by asynchronous execution. The diagram illustrates that in the synchronous case up to 1000s of GPUs have to wait until the analysis step finishes. In contrast, in an asynchronous workflow the GPUs/nodes don’t have to wait, significantly increasing the scalability of this heterogeneous use case.