scaling of adaptive sampling
for proteins
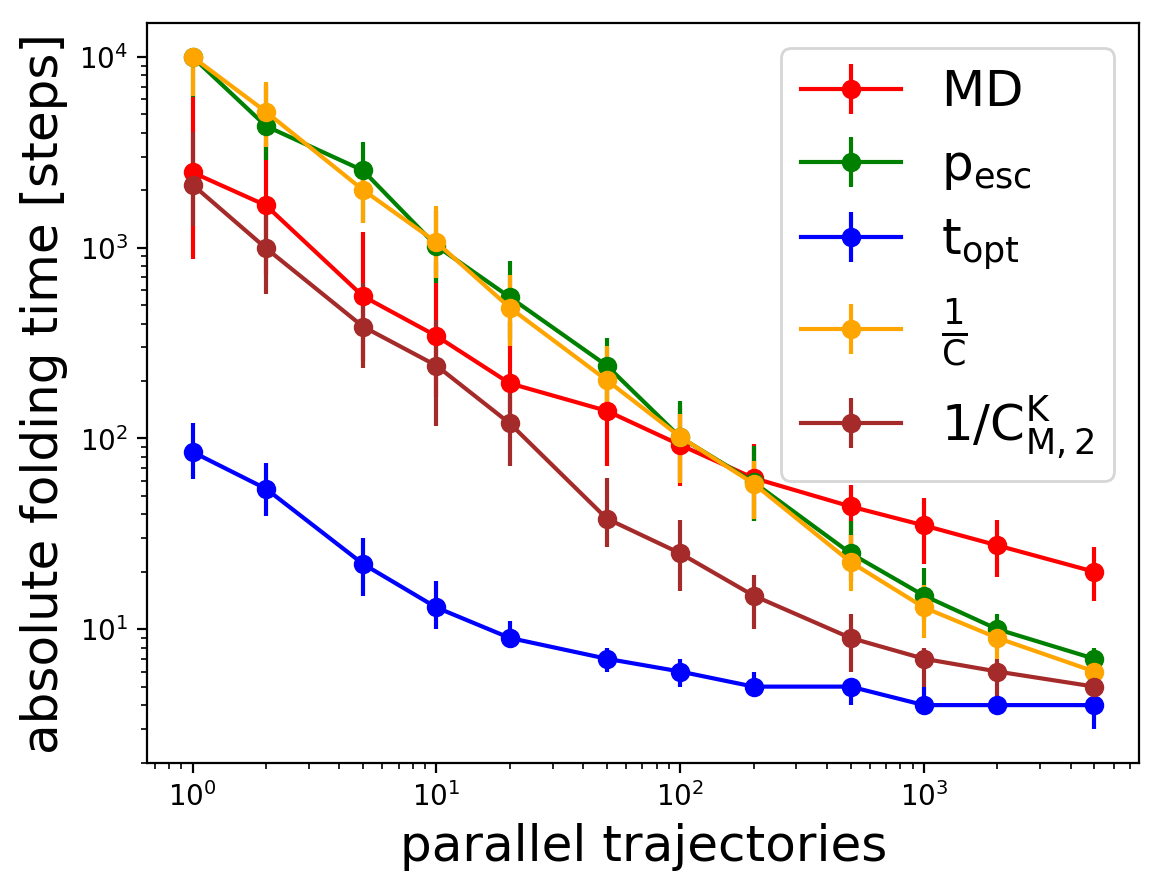
To understand adaptive sampling strategies better, the statistics of these simulations have to be improved, which is challenging due to the large amount of computational resources required to fold one single protein. Instead, a Markov Chain surrogate can be used. Here the scalability of adaptive sampling is investigated, indicating that different adaptive sampling strategies scale better (steeper curve) than molecular dynamics (MD) and the adaptive sampling scales to about 100-1000 concurrent molecular dynamics trajectories.
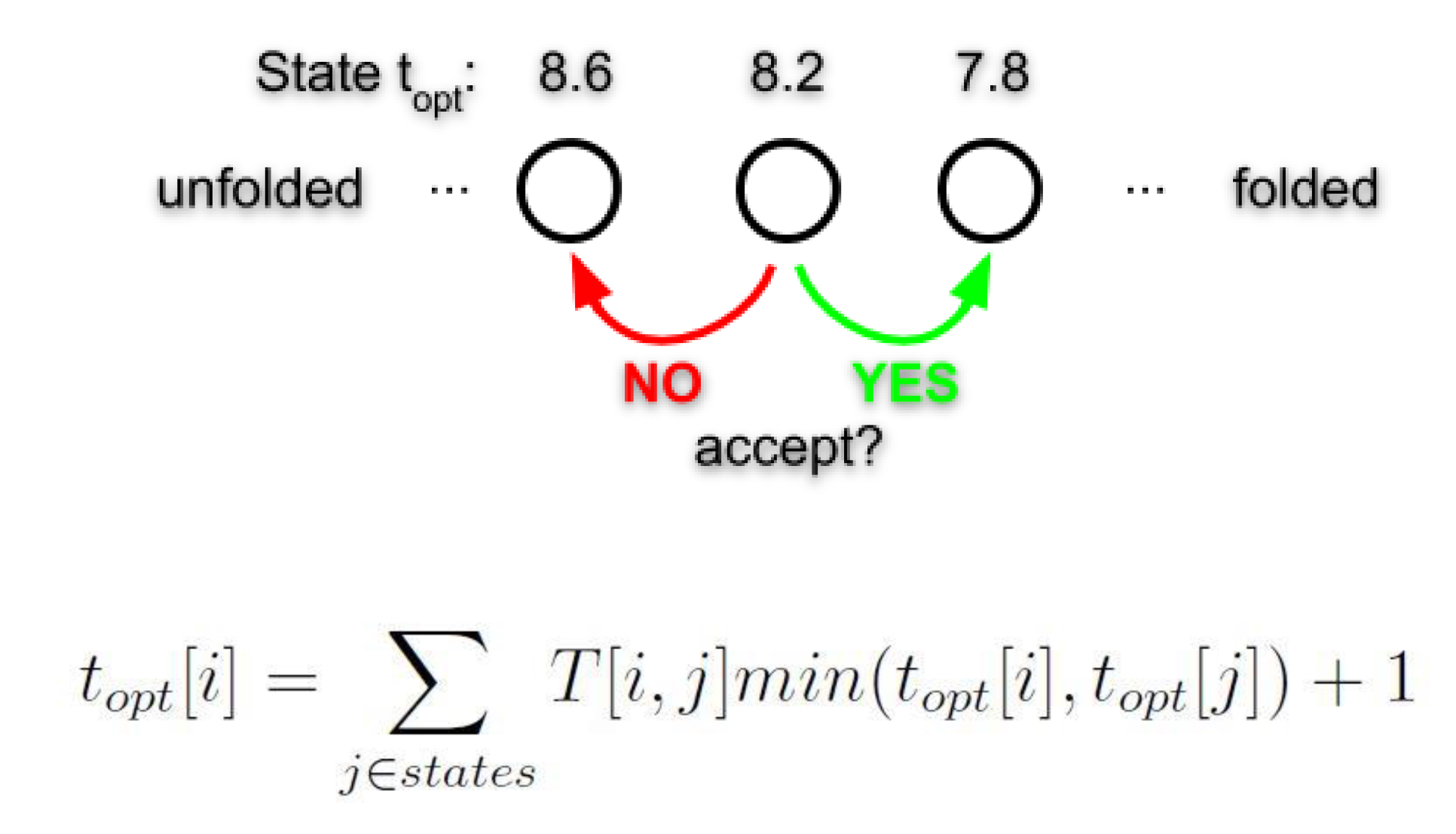
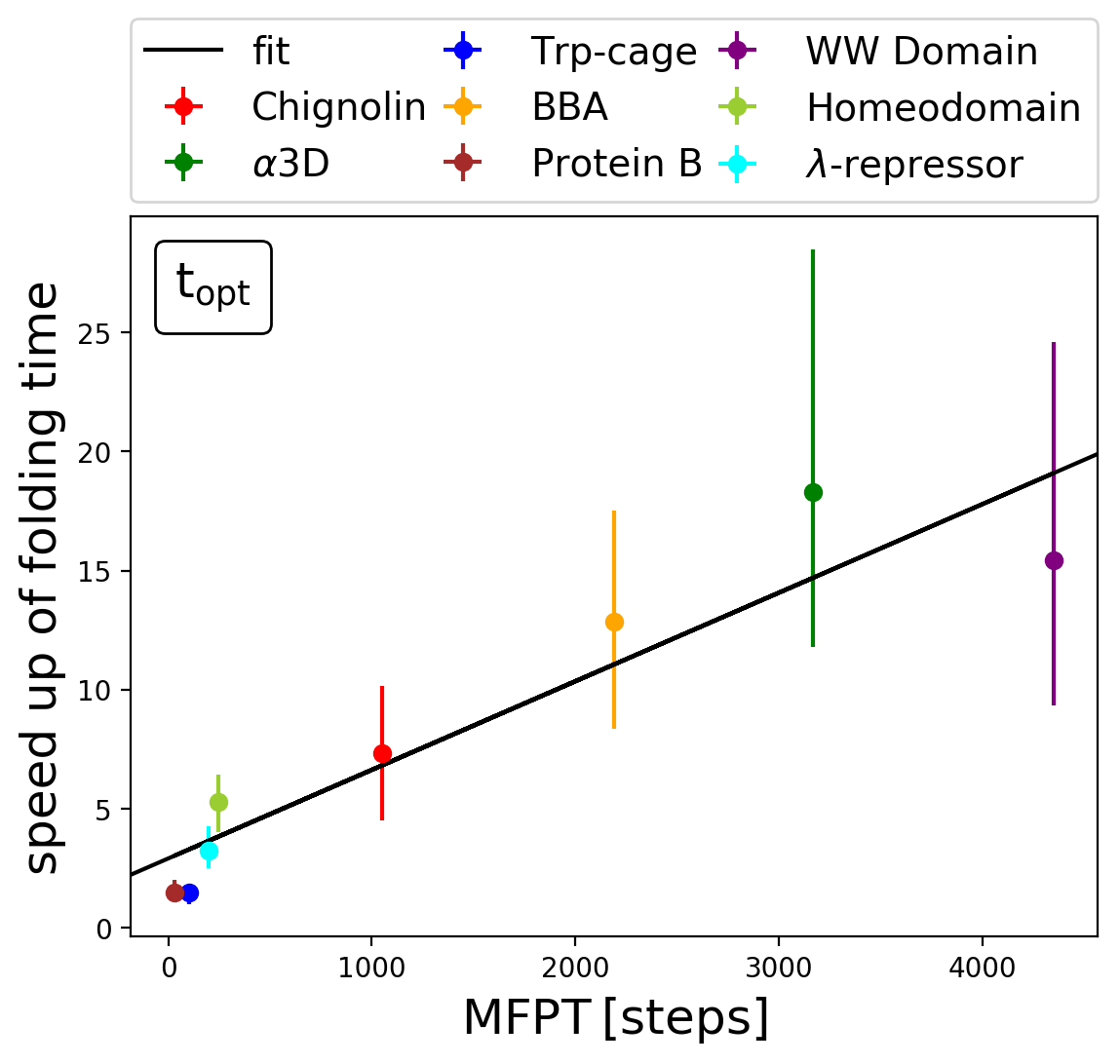
The maximum speed up with adaptive sampling compared to molecular dynamics was previously unclear, here I developed the first upper limit estimate with a greedy algorithm approach. The upper limit of the speed up with adaptive sampling reaches up to 20-fold for the small (<80 residues) proteins investigated. The speed up increases with the folding time, indicating that for larger proteins an even larger speed up is reachable.